2 General Information
The p53motifDB is a compendium of genomic locations in the human hg38 reference genome that contain recognizable DNA sequences matching the binding preferences for the transcription factor p53. Multiple types of genomic, epigenomic, and genome variation data were integrated with these locations in order to let researchers quickly generate hypotheses about novel activities of p53 or validate known behaviors.
2.1 Web access
The p53motif Database can be accessed via a docker-deployed Shiny App at https://p53motifdb.its.albany.edu/
2.2 Access to raw data
We maintain a Zenodo site containing all raw data tables, a downloadable Shiny app (usable via R, RStudio, or via Dockerfile), and a sqlite database version for advanced queries.
2.3 Feedback
Please let us know if you have any questions, comments, or would like additional datasets included in the next version of the p53motifDB by contacting masammons(at)albany.edu
2.4 General Use
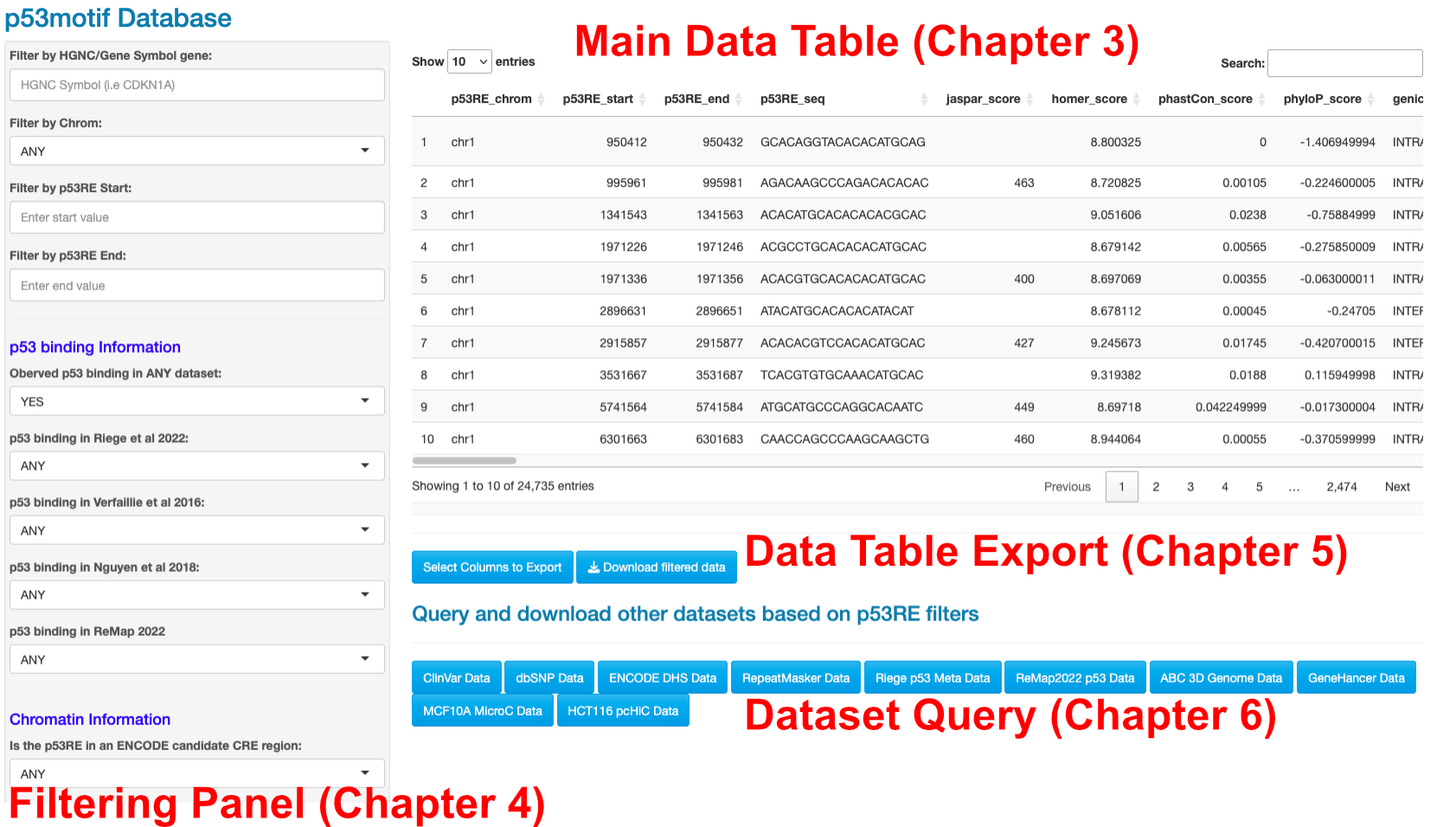
Figure 2.1: Layout of the p53motifDB Shiny App
The p53motifDB web app contains 4 components: the main data table (Chapter 3), the filtering panel (Chapter 4), the data table export section (Chapter 5), and the dataset query section (Chapter 6). These 4 components are used together to explore, filter, and export underlying data from the p53motifDB. Each component will be described in subsequent Chapters of this tutorial to help users navigate the web app and get the most out of the underlying data.