7 Database Summary Statistics
7.0.3 How many p53RE are found within 3D chromatin loops?
| ABC | GeneHancer | pCHiC - HCT116 | MicroC - MCF10A | |
|---|---|---|---|---|
| NO | 378073 | 324451 | 320978 | 317716 |
| YES | 34513 | 88135 | 91608 | 94870 |
7.0.6 How many p53RE are found within blacklisted regions of hg38?
| in blacklist? | number of p53RE |
|---|---|
| NO | 406423 |
| YES | 6163 |
7.0.7 How many p53RE are syntenic in the following assemblies?
| T2T | hg19 | MM10 | MM39 | |
|---|---|---|---|---|
| NO | 1375 | 1313 | 323962 | 323512 |
| YES | 411211 | 411273 | 88624 | 89074 |
7.0.8 How many p53RE found in different candidate Cis-Regulatory Elements (cCRE)?
| ccre_type | number of p53RE |
|---|---|
| CA | 12234 |
| CA-CTCF | 5398 |
| CA-H3K4me3 | 3363 |
| CA-TF | 1270 |
| PLS | 1426 |
| TF | 5918 |
| dELS | 69386 |
| pELS | 9302 |
| NA | 304289 |
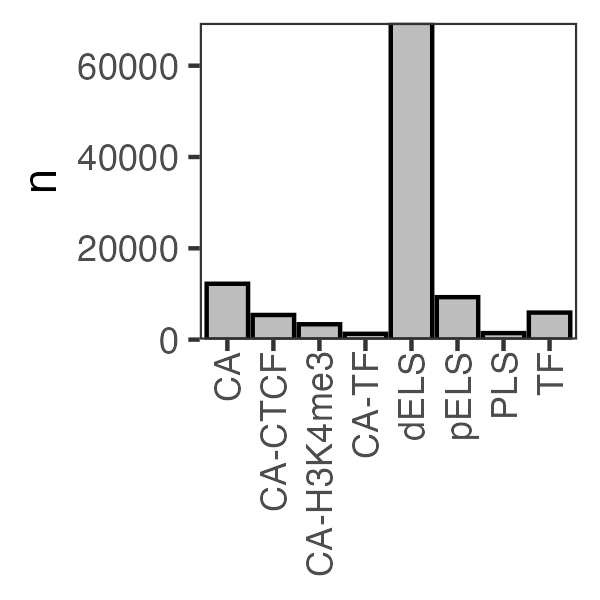
Figure 7.1: Number of p53RE in candidate Cis-Regulatory Elements (cCRE)
7.0.9 How many p53RE found in different chromHMM groupings?
| chromHMM_group | number of p53RE |
|---|---|
| acetylations | 36132 |
| bivalent promoters | 2489 |
| DNase | 698 |
| enhancers | 46017 |
| exon | 7787 |
| HET | 32230 |
| NA | 3525 |
| others | 20924 |
| polycomb repressed | 38782 |
| promoters | 3840 |
| quescient | 127791 |
| transcribed and enhancer | 12590 |
| transcription | 29199 |
| TSS | 463 |
| weak enhancers | 31349 |
| weak transcription | 15873 |
| znf | 2897 |
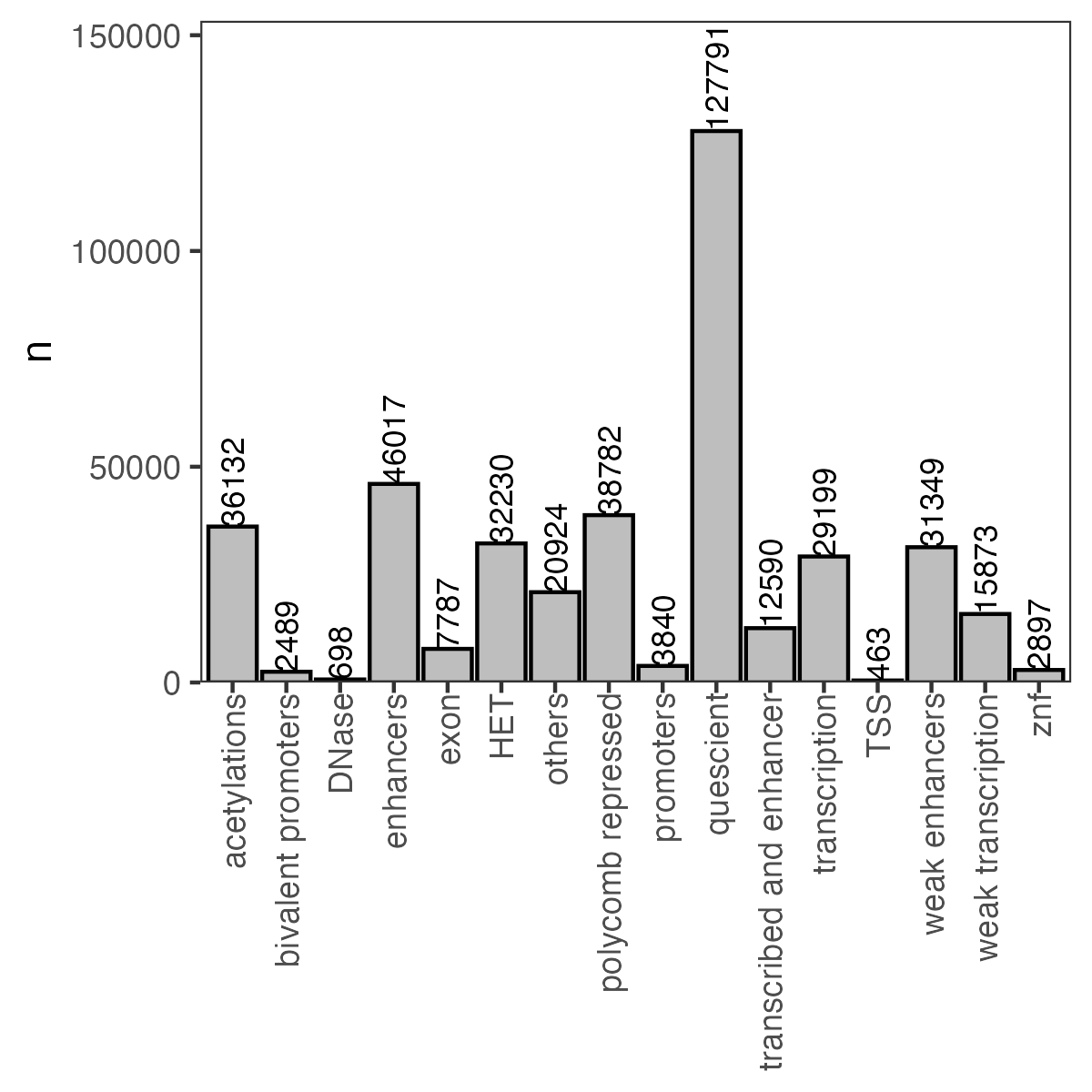
Figure 7.2: Number of p53RE in chromHMM Groupings
7.0.10 How many p53RE found in detailed chromHMM types?
| chromHMM mnemonic | number of p53RE |
|---|---|
| Acet1 | 4421 |
| Acet2 | 5322 |
| Acet3 | 13877 |
| Acet4 | 2155 |
| Acet5 | 3919 |
| Acet6 | 2011 |
| Acet7 | 1674 |
| Acet8 | 2753 |
| BivProm1 | 342 |
| BivProm2 | 443 |
| BivProm3 | 1260 |
| BivProm4 | 444 |
| DNase1 | 698 |
| EnhA1 | 941 |
| EnhA10 | 1667 |
| EnhA11 | 3091 |
| EnhA12 | 4372 |
| EnhA13 | 9294 |
| EnhA14 | 1625 |
| EnhA15 | 3928 |
| EnhA16 | 2318 |
| EnhA17 | 1901 |
| EnhA18 | 1897 |
| EnhA19 | 1386 |
| EnhA2 | 1306 |
| EnhA20 | 1056 |
| EnhA3 | 837 |
| EnhA4 | 1459 |
| EnhA5 | 2965 |
| EnhA6 | 2281 |
| EnhA7 | 1870 |
| EnhA8 | 1073 |
| EnhA9 | 750 |
| EnhWk1 | 5446 |
| EnhWk2 | 1754 |
| EnhWk3 | 3449 |
| EnhWk4 | 7891 |
| EnhWk5 | 3923 |
| EnhWk6 | 2528 |
| EnhWk7 | 1977 |
| EnhWk8 | 4381 |
| GapArtf1 | 20850 |
| GapArtf2 | 54 |
| GapArtf3 | 20 |
| HET1 | 4392 |
| HET2 | 3510 |
| HET3 | 6202 |
| HET4 | 1987 |
| HET5 | 1547 |
| HET6 | 2864 |
| HET7 | 4926 |
| HET8 | 2531 |
| HET9 | 4271 |
| PromF1 | 1085 |
| PromF2 | 585 |
| PromF3 | 453 |
| PromF4 | 411 |
| PromF5 | 364 |
| PromF6 | 393 |
| PromF7 | 549 |
| Quies1 | 36125 |
| Quies2 | 14307 |
| Quies3 | 49915 |
| Quies4 | 20321 |
| Quies5 | 7123 |
| ReprPC1 | 699 |
| ReprPC2 | 1290 |
| ReprPC3 | 4372 |
| ReprPC4 | 15527 |
| ReprPC5 | 3247 |
| ReprPC6 | 6731 |
| ReprPC7 | 3084 |
| ReprPC8 | 2403 |
| ReprPC9 | 1429 |
| TSS1 | 245 |
| TSS2 | 218 |
| Tx1 | 3235 |
| Tx2 | 6519 |
| Tx3 | 2870 |
| Tx4 | 1958 |
| Tx5 | 4958 |
| Tx6 | 3589 |
| Tx7 | 3082 |
| Tx8 | 2988 |
| TxEnh1 | 1542 |
| TxEnh2 | 1569 |
| TxEnh3 | 1376 |
| TxEnh4 | 1655 |
| TxEnh5 | 2691 |
| TxEnh6 | 884 |
| TxEnh7 | 1540 |
| TxEnh8 | 1333 |
| TxEx1 | 1227 |
| TxEx2 | 2650 |
| TxEx3 | 3400 |
| TxEx4 | 510 |
| TxWk1 | 11628 |
| TxWk2 | 4245 |
| znf1 | 2025 |
| znf2 | 872 |
| NA | 3525 |
7.0.11 p53RE location by gene location
| Gene Location | number of p53RE |
|---|---|
| INTERGENIC | 158859 |
| INTRAGENIC | 253727 |
7.0.12 p53RE location by chromosome
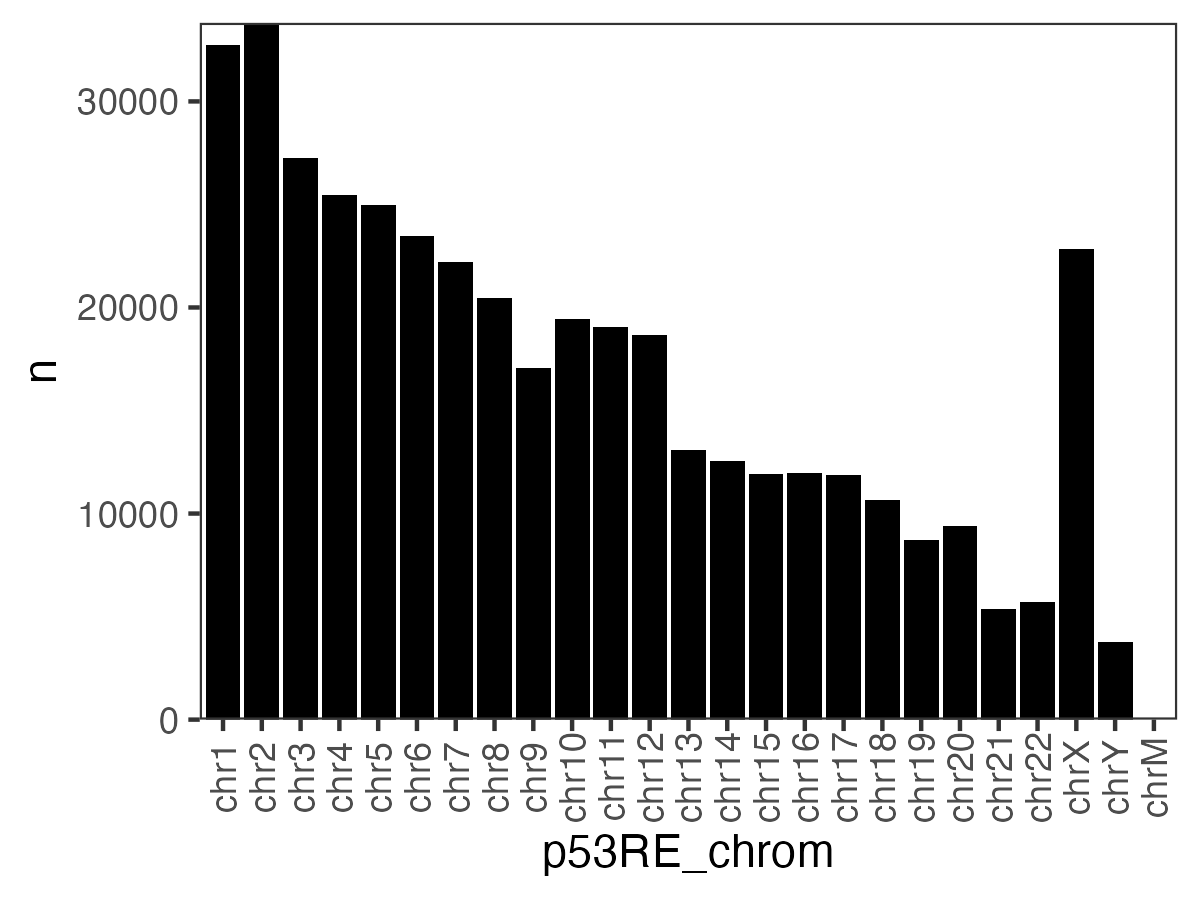
Figure 7.3: Location of p53RE on each chromosome
| chromosome | number of p53RE |
|---|---|
| chr1 | 32725 |
| chr2 | 33813 |
| chr3 | 27249 |
| chr4 | 25479 |
| chr5 | 24977 |
| chr6 | 23486 |
| chr7 | 22207 |
| chr8 | 20460 |
| chr9 | 17044 |
| chr10 | 19444 |
| chr11 | 19053 |
| chr12 | 18671 |
| chr13 | 13107 |
| chr14 | 12562 |
| chr15 | 11910 |
| chr16 | 11988 |
| chr17 | 11884 |
| chr18 | 10638 |
| chr19 | 8741 |
| chr20 | 9397 |
| chr21 | 5381 |
| chr22 | 5725 |
| chrM | 2 |
| chrX | 22862 |
| chrY | 3781 |